Volumetric data rendering with Neuroglancer
Deborah Schmidt
Helmholtz Imaging | MDC Berlin
Sep 25, 2024
Introduction
Main Features:
- Voxel-based rendering: Efficiently visualize large 3D datasets by streaming the data directly into the browser.
- Support for multiple data formats: Neuroglancer works with a variety of formats like ZARR and OME-ZARR for volumetric images, as well as annotation formats.
- Interactive, shareable views: Customize and share views by copying the URL directly from the Neuroglancer interface.
Available datasets
Popular datasets using Neuroglancer
- MICrONS Explorer: A large-scale dataset from the MICrONS Project, providing high-resolution volumetric reconstructions of a mouse brain.
- FlyEM Hemibrain: This dataset offers a detailed 3D reconstruction of the Drosophila melanogaster brain at nanometer resolution.
- “H01” Dataset: 1.4 Petabyte for one cubic millimeter of the human brain, released by the Harvard University and the Connectomics at Google team.
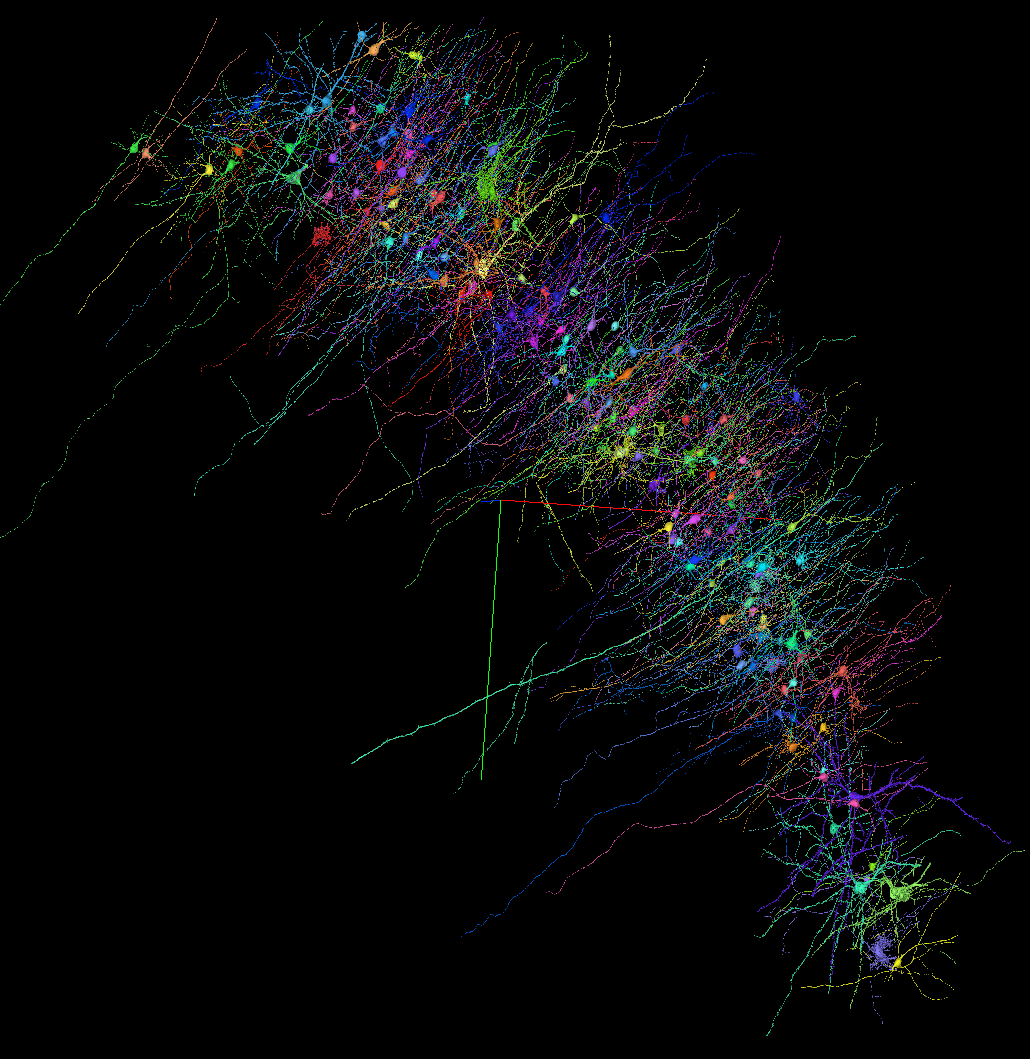
Screenshot from the H01 Dataset (link)
Available datasets
Helmholtz Imaging collaboration use case
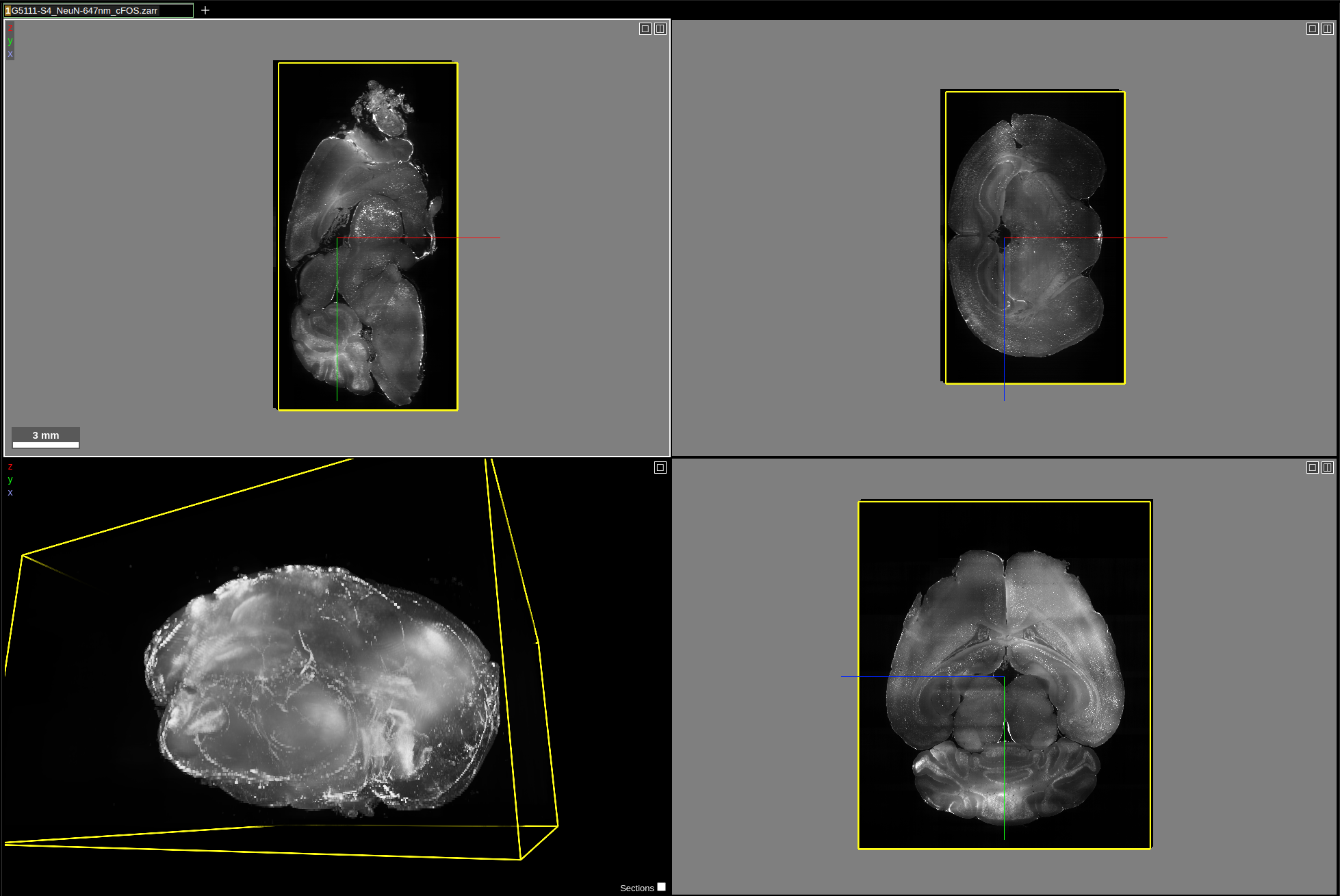
Dataset requirements
Supported Data Types
- Neuroglancer precomputed format
- N5
- Zarr v2/v3 (including OME-ZARR)
- Python in-memory volumes (with automatic mesh generation)
- BOSS , DVID, Render, Single NIfTI files, Deep Zoom images
Data preparation
Volumetric images
Conversion Steps:
- Step 1: Convert the TIFF slices into ZARR format, which supports chunked, compressed storage.
- Step 2: Convert the ZARR files into OME-ZARR format, which includes metadata for biological imaging data and supports better integration with visualization tools like Neuroglancer.
Data preparation
Annotations
Conversion Steps:
- Step 1: Prepare a list of points or annotations that describe features in your dataset (e.g., 3D coordinates for cell locations).
- Step 2: Convert the list of points into CloudVolume annotations.
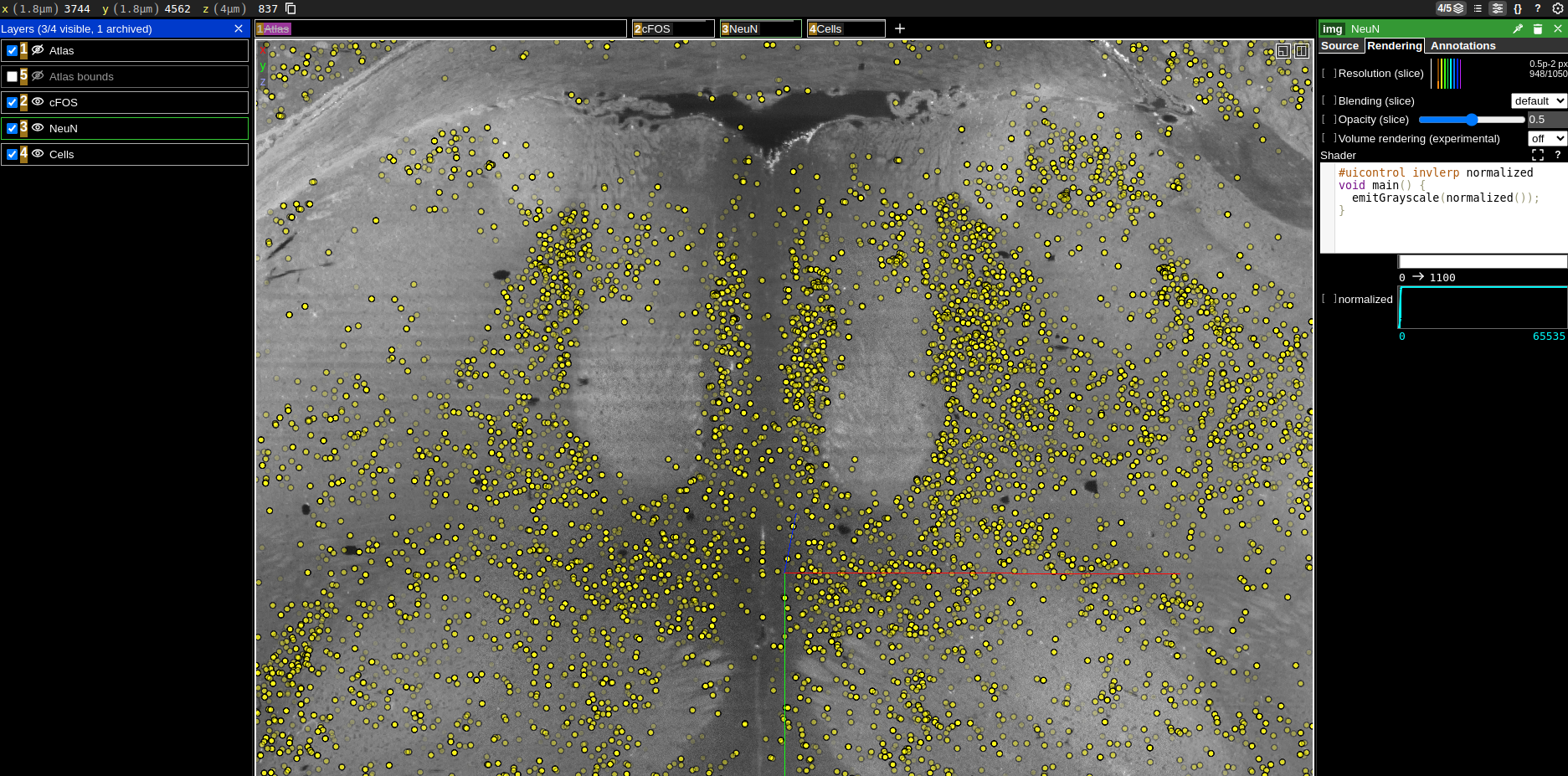
Streaming data locally
With Python:
- Step 1: Use Python’s built-in HTTP server to serve the data directory:
python3 -m http.server - Step 2: Open the Neuroglancer demo page and enter the local URL of your data (e.g.,
zarr://http://localhost:8000/my-dataset.ome.zarr).
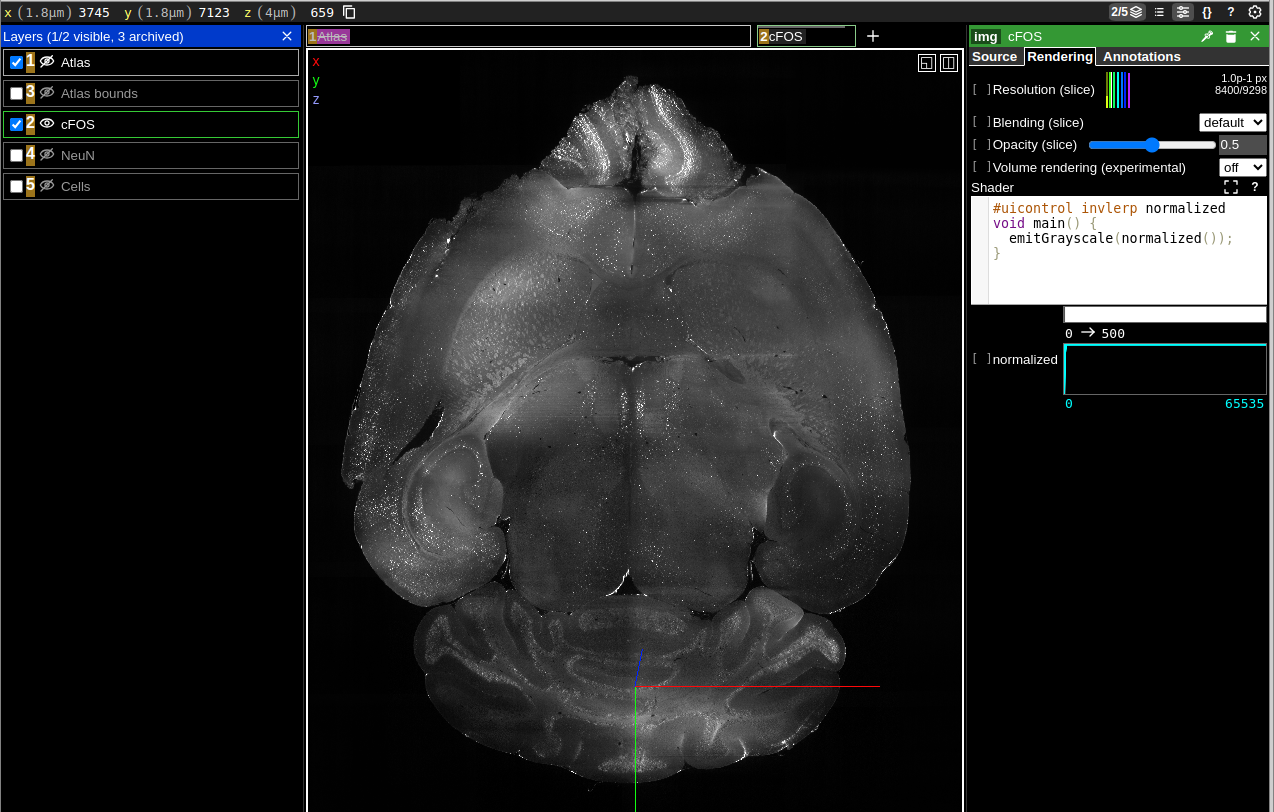
Sharing views
Example of JSON in URL:
- Copy the URL from your browser, and you’ll notice that it contains encoded parameters for layers, views, and more.
- The JSON configuration embedded in the URL can be customized to adjust the view programmatically.
{
"dimensions": {...},
"position": [
3744.089599609375,
4562.4326171875,
837.0703125
],
"layers": [
{
"type": "image",
"source": "zarr://https://hifis-storage.desy.de:2443/Helmholtz/HIP/collaborations/2405_MDC_Treier/public/G5111-S4/647nm_cFOS",
"tab": "rendering",
"shaderControls": {
"normalized": {
"range": [
0,
500
]
}
},
"crossSectionRenderScale": 0.47742080195520836,
"name": "cFOS"
}
{
"type": "annotation",
"source": "precomputed://https://hifis-storage.desy.de:2443/Helmholtz/HIP/collaborations/2405_MDC_Treier/public/G5111-S4/cells",
"tab": "source",
"name": "Cells"
}
],
"selectedLayer": {
"visible": true,
"layer": "cFOS"
}
}
Programmatically generate URL
Link to the full notebook showing how to generate a Neuroglancer URL programmatically:
Data hosting for Neuroglancer
- Local Hosting: Serve data from your own machine or institution’s servers.
- AWS S3 or Google Cloud Storage: Store large datasets and stream them to Neuroglancer from the cloud.
- Scientific Data Services: e.g. BioImage Archive
- Do you know other compatible hosting places? Let us know!
Data hosting for Neuroglancer
Helmholtz storage compatible to Neuroglancer
Collaboration between Helmholtz Imaging and HIFIS at DESY, with the support of the Jülich Cluster.


- dCache / InfiniteSpace: Secure, scalable storage solution for Helmholtz researchers to host and stream datasets. The data hosted on the storage can be shared publicly.
Steps:
- Authenticate yourself to access the storage using AAI.
- Upload the data, i.e. using
rclonecommand line tool or theRclone BrowserGUI
Data hosting for Neuroglancer
Helmholtz Neuroglancer Instance
- Helmholtz Imaging Neuroglancer instance: A dedicated Neuroglancer instance for the Helmholtz community.
Steps:
- In https://hifis-storage.desy.de, right click on the dataset you want to stream and click “Get WebDAV link”
- Replace the
2880port with2443 - Open https://neuroglancer.helmholtz-imaging.de
- Add your dataset using the modified URL